6 iPhylo CLI
The iPhylo Command-Line Interface (CLI) is an extension of the web-based iPhylo Tree. It operates offline and comes with integrated databases, which can be deployed either locally or on high-performance computing clusters for enhanced performance.
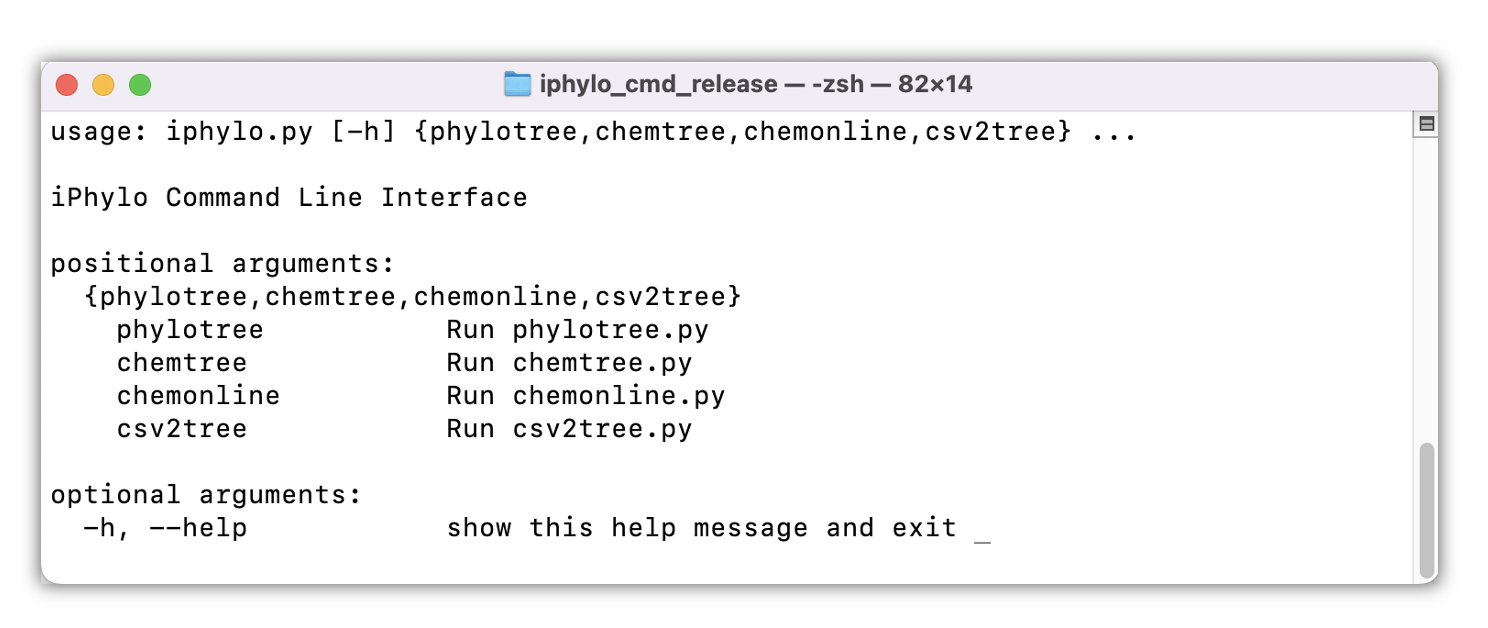
Importantly, the iPhylo CLI can also construct customized taxonomic trees based on a user-defined hierarchical database so the applications of the iPhylo suite can be extended beyond biological and chemical classifications.
The GitHub repository for iPhylo CLI can be accessed via https://github.com/ARise-fox/iPhylo-CLI/
The iPhylo CLI includes four modules: