4 Input & Output
4.1 Input
4.1.1 Input elements
You can use the following element types, or any combination of them. Biological elements should be separated by commas or newlines. Chemical elements should be separated by newlines to prevent potential misinterpretation of names due to commas.
4.1.2 Biological
- Organism names: The names are flexible as long as they are indexed in the NCBI taxonomic database, such as “scientific name”, “synonym”, or “common name”. For example, “human” can serve as the common name for “Homo sapiens”. All leaf nodes within the iPhylo trees will be their corresponding scientific name. You can see how name changed in the output text file. For instance:
honey bee -> Apis mellifera
house mouse -> Mus musculus
human -> Homo sapiensTaxonomy identifiers (taxid)
Subtree: Follows the format “xxx|subtree”, see details in Sub-tree.
4.1.3 Chemical
- InChIKey: The InChIKey is a fixed length (27 character) condensed digital representation of the InChI that is not designed to be human-understandable.
We strongly recommend using the InChIKey to represent chemicals in the chemical tree online module, which utilizes the chemonline.py from the ClassyFire API in iPhylo CLI, due to its unique and concise nature.
For example:
VXRWAWLEFDIKKA-NGVUZZMQSA-N
JWUMPSYUUYOAEP-GLYJLXGFSA-N
PLYVHUNKJZPXRH-RCNNPCQASA-N- isomeric SMILES The simplified molecular-input line-entry system (SMILES) is a specification in the form of a line notation for describing the structure of chemical species using short ASCII strings.
This format is highly recommended for use in the chemical tree online module, which employs the NPonline.py from the NPClassifier API in iPhylo CLI.
For example:
O=C(NC(COC1OC(CO)C(OC2OC(CO)C(O)C(O)C2O)C(O)C1O)C(O)C=CCCC=CCCC=CCCCCCC)CCCCCCCCC=CCC=CCC=CCC=CCC- InChI The International Chemical Identifier (InChI) is a textual identifier for chemical substances, designed to be structure-based, strictly unique, and non-proprietary.
For example:
InChI=1S/C23H25N5O5/c1-30-18-11-14-15(12-19(18)31-2)25-23(26-21(14)24)28-9-7-27(8-10-28)22(29)20-13-32-16-5-3-4-6-17(16)33-20/h3-6,11-12,20H,7-10,13H2,1-2H3,(H2,24,25,26)/t20-/m1/s1- Subtree: Follows the format “xxx|subtree”, see details in Sub-tree.
4.2 Output
4.2.1 Tree file
iPhylo can generate trees in the following formats: Newick, Nexus, and PhyloXML.
- Newick Format:
Description: Newick, also known as New Hampshire or New Hampshire Extended format, is a simple and widely used text-based format for representing phylogenetic trees. It expresses tree structures using nested parentheses and commas.
Example:
(A:0.1,B:0.2,(C:0.3,D:0.4):0.5);Usage: It’s commonly used for representing hierarchical relationships in evolutionary biology and bioinformatics.
- Nexus Format:
Description: Nexus is a versatile file format that can store various types of biological data, including phylogenetic trees. It allows the inclusion of metadata, DNA/protein sequences, and more. It is both human-readable and writable.
Example:
#NEXUS Begin trees; Tree myTree = (A,B,(C,D)); End;Usage: Nexus is often used in phylogenetics, systematics, and evolutionary biology due to its flexibility.
- PhyloXML:
Description: PhyloXML is an XML-based format designed to store and exchange phylogenetic trees and associated data. It supports a variety of information, including node labels, branch lengths, and annotations.
Example:
Usage: PhyloXML is suitable for storing and sharing complex phylogenetic data, often used in bioinformatics.
- Phylip Format:
Description: The Phylip (PHYLogeny Inference Package) format is a simple, line-based format developed for use with the Phylip software package. It is used to represent both sequence data and phylogenetic trees.
Example:
4 A 0.0 0.1 0.2 B 0.1 0.0 0.3 C 0.2 0.3 0.0 D 0.3 0.2 0.1Usage: Phylip format is commonly used for input and output in phylogenetic analysis software and tools.
These formats cater to different needs, from simple tree representations to more complex structures with additional metadata. The choice of format often depends on the specific requirements of the analysis or tool being used.
4.2.2 Tree structure visualizations
ASCII Tree: Simple pretty-printing of your tree structures using pure ASCII characters for drawing branches and edges.
For instance:
_____ ____ _____ ____ _____ _____ s__Drosophila_melanogaster
|
_____ ____| _____ ____ _____ _____ s__Mus_musculus
| | ____|
| |_____| |_____ ____ _____ _____ s__Homo_sapiens
______| |
| |____ _____ ____ _____ _____ s__Gallus_gallus
|
|_____ ____ _____ ____ _____ ____ _____ _____ s__Escherichia_coli
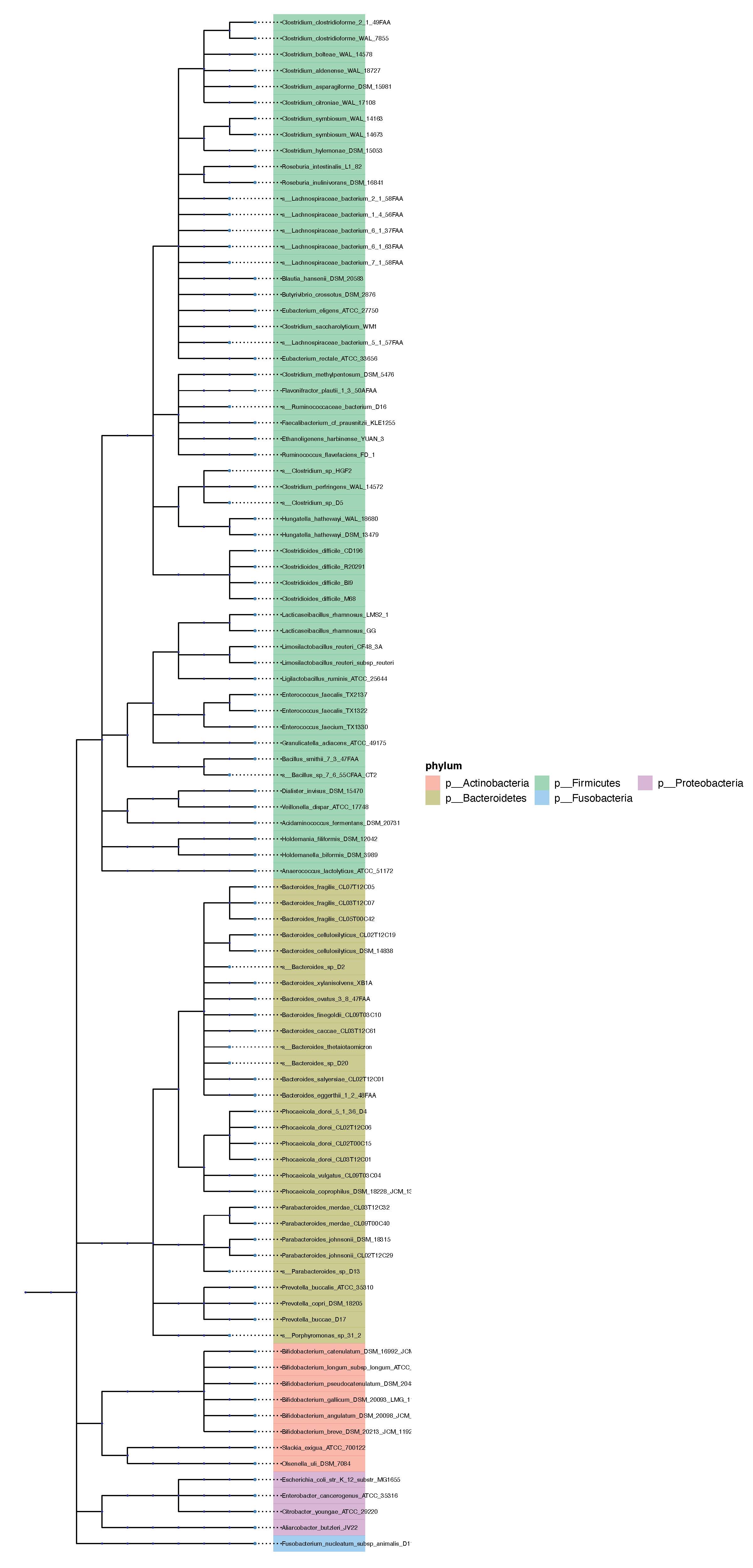
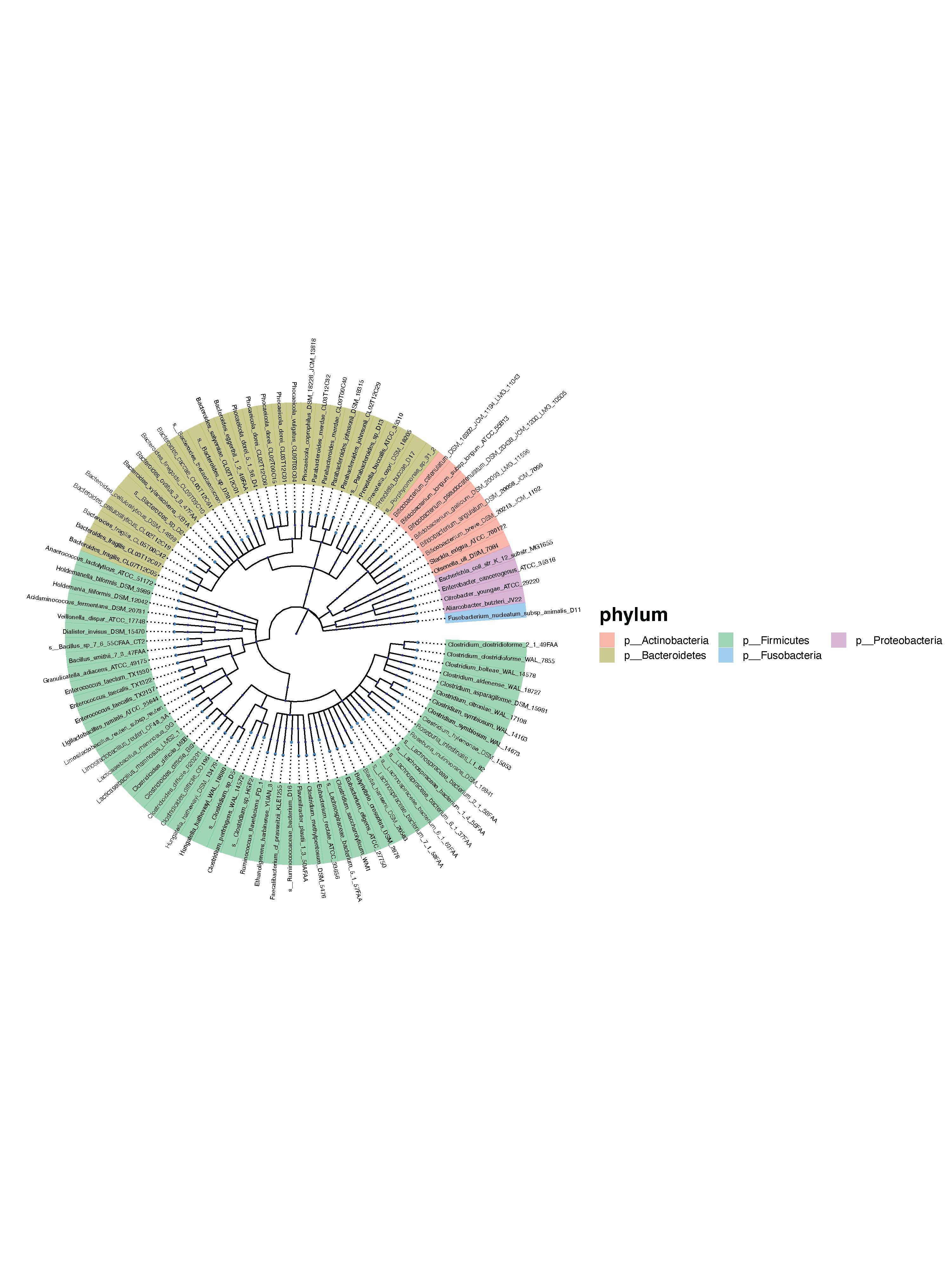
Simple tree plot: The phylograph in PDF format, with clades colored by phyla or superclass. Plots are drawn using ggtree
For instance:
- Rectangular Plot
- Circular Plot
4.2.3 Additional files
When running iPhylo, we generate some other files to help you understand the tree better:
iphylo_tree_items.csv,ichem_tree_items.csv
These spreadsheet enlist the taxID (id for chemicals), scientific name (name, InChI, InChIKey, SMILES for chemicals) and lineage for the taxa you input.
iphylo_tree_items_for_anno.csv
Similar to iphylo_tree_items.csv, but tailored for iPhylo Visual, serves as an annotation for basic visualization of tree leaves. It can be directly uploaded to iPhylo Visual for enhanced visualization and annotation.
no_match_result.txt
This file is used to alert you when you have input taxIDs and names that are not in the NCBI taxonomy database, or when you have input a name that corresponds to more than one taxID. Note that the taxID is the unique identifier for NCBI taxonomy database entries.
In the case of chemical trees, this file is used to indicate which compounds are not in the iPhylo database or are missing classification information. Our database mainly contains functional compounds, if you want more more compounds information, try our iPhylo CLI chemoline module.