1 Introduction
1.1 Network
In mathematics, “networks” are often referred to as “graphs”, and the mathematical field of graph research is called “graph theory”.
The basic elements in a network graph are nodes and edges. When constructing a network graph, the objects are called “nodes” (vertices or nodes), and they are usually drawn as points; the connections between nodes are called “edges” (edges or links), and they are usually drawn as lines between points.
We can be divide networks into directed and undirected networks, weighted and unweighted networks according to the edges.
1.2 Network in omics
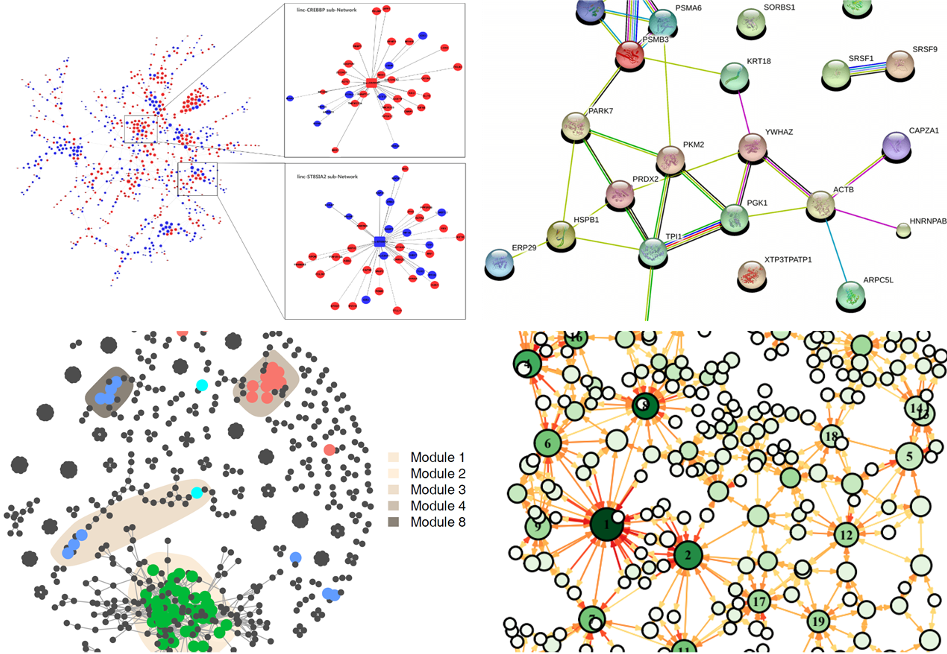
Network can represent various systems in the real world,and it have many applications in biological research, especially in systems biology: gene expression regulatory networks, metabolic networks, ecosystem space networks, microbial co-occurrence networks, protein interaction networks, etc (Figure 1.1).

WGCNA
Weighted Gene Co-expression Network Analysis (WGCNA) is a widely used method to construct gene, protein, or metabolite co-expression networks. It is a systems biology method for describing the correlation patterns among genes across microarray samples.
WGCNA can be used to find clusters (modules) of highly correlated genes, to summarize such clusters using the module eigengene or an intramodular hub gene, to relate modules to one another and to external sample traits (using eigengene network methodology), and to calculate module membership measures. Correlation networks are first constructed, and then the adjacency matrix is transformed into a topological overlap matrix (TOM) to further enhance the network properties.
Co-occurrence
Co-occurrence networks are used to describe the co-occurrence patterns of different species in a community. The nodes in the network represent species, and the edges represent the co-occurrence relationships between species. The co-occurrence network can be used to study the interaction between species in the community, the stability of the community, and the response of the community to environmental changes.
PPI
Protein-protein interaction (PPI) networks are used to describe the interactions between proteins. The nodes in the network represent proteins, and the edges represent the interactions between proteins. PPI networks can be used to study the functions of proteins, the pathways in which proteins are involved, and the relationships between proteins.
Metabolic
Metabolic networks are used to describe the metabolic pathways in cells. The nodes in the network represent metabolites, and the edges represent the metabolic reactions between metabolites. Metabolic networks can be used to study the metabolic pathways in cells, the relationships between metabolites, and the regulation of metabolic pathways.
1.3 Software
There are many software packages for network analysis, including R, Python, Pajek, Cytoscape, and Gephi.
R: igraph (https://igraph.org/), network
Python: networkx (https://pypi.org/project/networkx/)
Cytoscape (https://cytoscape.org/)
Gephi (https://gephi.org/)