MetaNet: Network analysis for multi-omics
The HTML documentation of the latest version is available at Github page.
Installation
You can install the released version of MetaNet from CRAN with:
install.packages("MetaNet")You can install the development version of MetaNet from GitHub with:
# install.packages("devtools")
devtools::install_github("Asa12138/MetaNet")Workflow overview

Figure 1. Overview of the MetaNet workflow and its high-efficiency computation. (A) Functional modules of MetaNet, visualized using MetaNet itself. (B) Detailed workflow of MetaNet. Green boxes indicate data objects, red boxes represent MetaNet-specific objects, and gray boxes denote core functions. (C) MetaNet logo and its available code repositories and platforms. (D) Line plots comparing memory usage and runtime for correlation-based network construction across different R packages. Error bars represent standard deviation (SD). (E) Line plots showing MetaNet’s performance on increasingly larger datasets in terms of memory usage and runtime. Error bars represent SD.
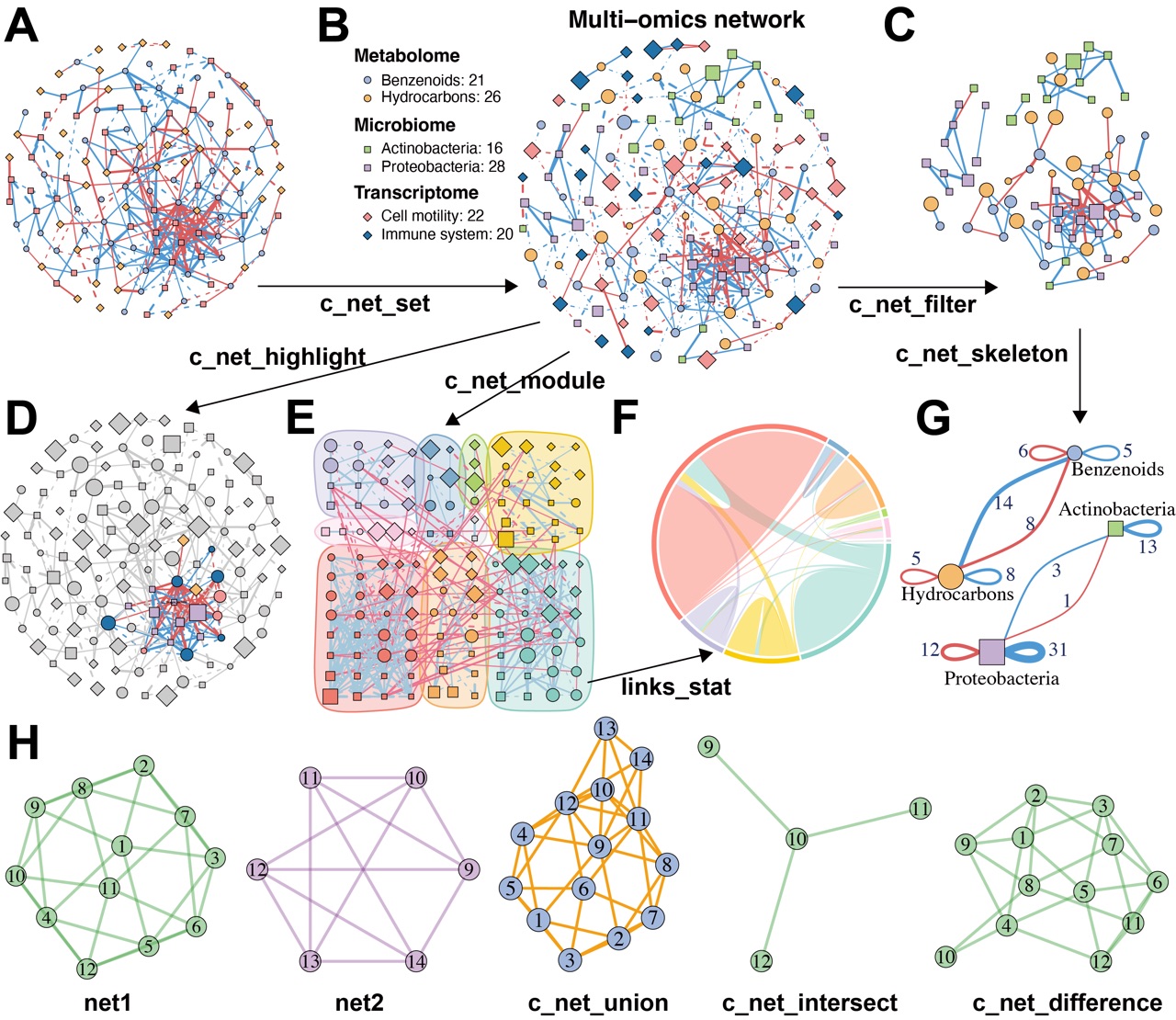
Figure 2. MetaNet supports flexible and intuitive network manipulation. (A) Initial multi-omics network constructed without annotations. (B) Annotated multi-omics network using the “c_net_set” function. Node shape indicates omics type, color represents omics subtypes, size denotes average abundance, edge color indicates positive or negative correlation, edge type distinguishes intra- and inter-omics connections, and edge width reflects the absolute value of the correlation coefficient. (C) Subnetwork filtered from intra-omics interactions between the Microbiome and Metabolome layers using “c_net_filter”. (D) Highlighted nodes centered on “s__Dongia_mobilis” and its neighbors using “c_net_highlight”. (E) Community detection and modular visualization using “c_net_module”. (F) Chord diagram displaying the proportion of edges between modules. (G) Skeleton network across omics subtypes at a grouped level using “c_net_skeleton”. (H) Operations among networks: “c_net_union” merges net1 and net2, “c_net_intersect” extracts shared nodes and edges, and “c_net_difference” isolates net1-specific nodes and edges. All networks shown are based on simulated data and are for illustrative purposes only.
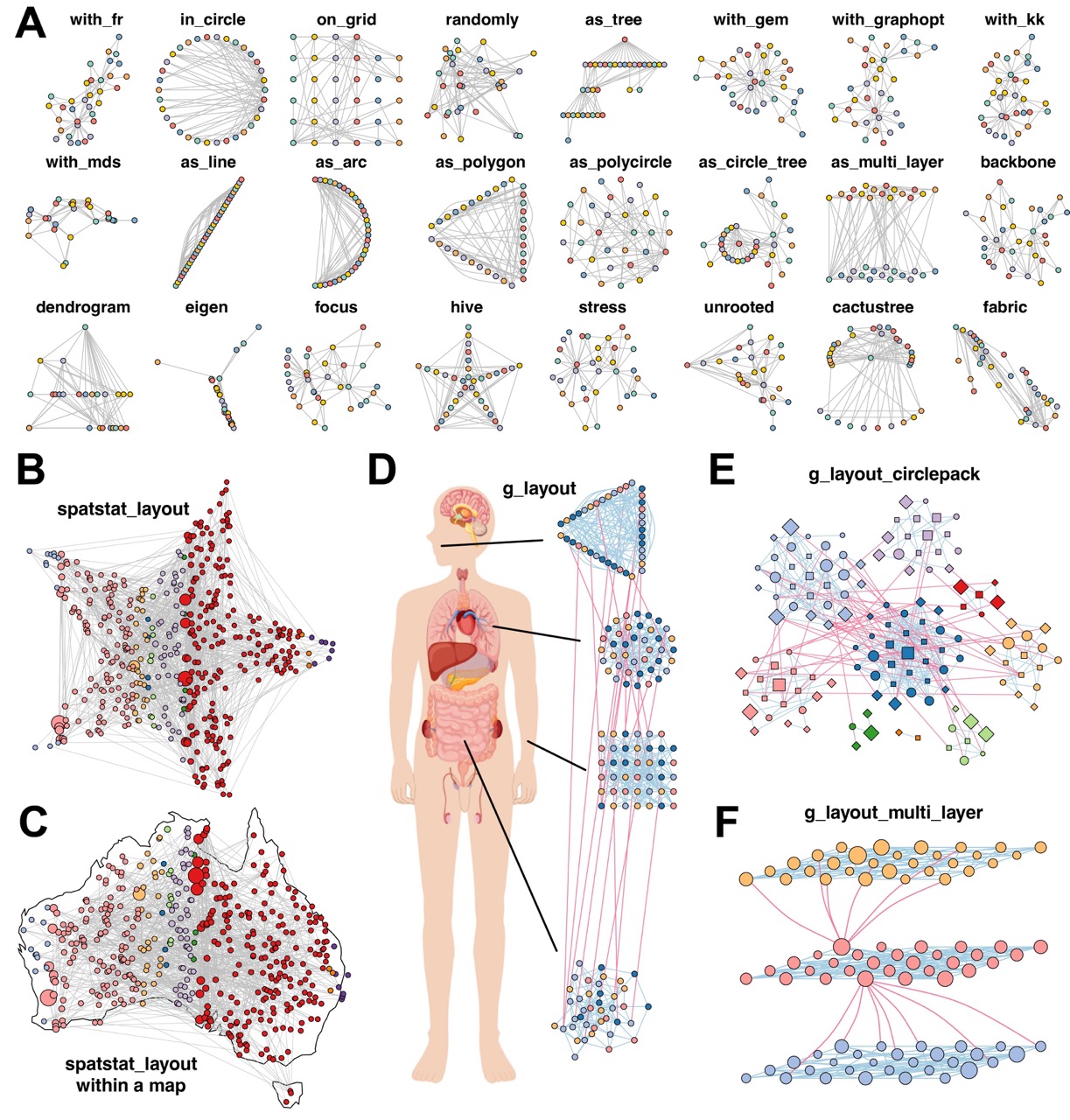
Figure 3. MetaNet enables diverse and powerful network layout strategies. (A) Application of 24 out of more than 40 built-in layout algorithms from “c_net_layout” on the Zachary Karate Club network. (B) Layout generated within a star using “spatstat_layout”. (C) Layout applied within the map of Australia using “spatstat_layout”. (D) Grouped network layout consisting of four subgroups arranged with “with_fr()”, “on_grid()”, “as_polycircle(3)”, and “as_polygon(3)” within a human-body schematic. All visualization elements were rendered entirely in MetaNet without manual adjustment. (E) Modular network visualized using “g_layout_circlepack”. (F) Three-layer modular structure visualized using “g_layout_multi_layer”. All networks shown are based on simulated data and carry no biological interpretation.
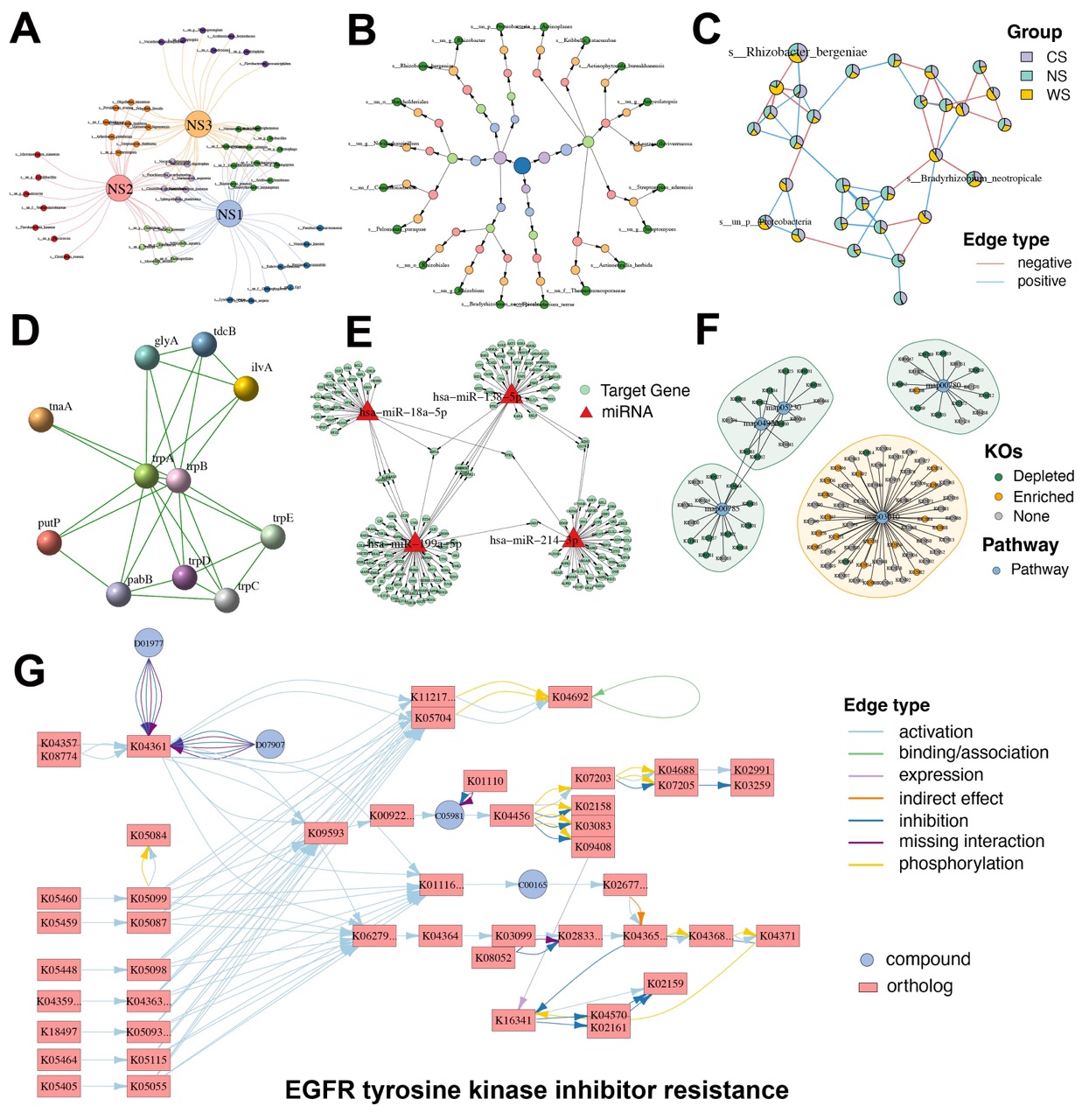
Figure 4. Diverse specialized network visualizations by MetaNet. (A) Venn-style network: Large nodes represent groups, while smaller nodes denote individual elements within each group, enabling visualization of shared and unique components. (B) Hierarchical tree network: Nodes are organized based on classification hierarchy. Node color corresponds to the taxonomic or categorical level. (C) Pie-node network: Each node is displayed as a pie chart, where slice colors indicate relative abundance across different groups. (D) Protein–protein interaction (PPI) network: Extracted from the STRING database, showing experimentally validated and predicted molecular interactions among proteins. (E) miRNA–gene regulatory network: Sourced from the miRTarBase database, illustrating experimentally supported regulatory relationships between miRNAs and their target genes. (F) KEGG KO–pathway association network: The network shows KEGG orthologs (KOs) involved in selected biological pathways. Small nodes represent KOs, and large nodes represent pathways. KO nodes are colored by their expression trend. Shaded regions surrounding pathways indicate whether the pathway is globally up-regulated (orange) or down-regulated (green). (G) KEGG pathway-specific network: Network representation of the “EGFR tyrosine kinase inhibitor resistance” pathway. Rectangular nodes denote KEGG orthologs, circular nodes indicate compounds, and edge colors reflect interaction types.
Citation
Please cite:
- Peng, C. et al. MetaNet: a scalable and integrated tool for reproducible omics network analysis. 2025.06.26.661636 Preprint at https://doi.org/10.1101/2025.06.26.661636 (2025).
Need helps?
If you have questions/issues, please visit MetaNet homepage first. Your problems are mostly documented. If you think you found a bug, please post on github issue.