Get RMT threshold for a correlation matrix
Get RMT threshold for a correlation matrix roughly
RMT_threshold(
occor.r,
out_dir,
min_threshold = 0.5,
max_threshold = 0.8,
step = 0.02,
plot = FALSE,
gif = FALSE,
verbose = FALSE
)
rmt(occor.r, min_threshold = 0.5, max_threshold = 0.85, step = 0.01)Arguments
Value
a r-threshold
recommend threshold
References
J. Zhou, Y. Deng, FALSE. Luo, Z. He, Q. Tu, X. Zhi, (2010) Functional Molecular Ecological Networks, doi:10.1128/mBio.00169-10. https://matstat.org/content_en/RMT/RMThreshold_Intro.pdf
Examples
# \donttest{
data(otutab, package = "pcutils")
t(otutab) -> totu
c_net_calculate(totu) -> corr
rmt(corr)
#> Calculating: 0.5
#> Calculating: 0.6
#> Calculating: 0.65
#> Calculating: 0.7
#> Calculating: 0.75
#> Calculating: 0.8
#> Calculating: 0.85
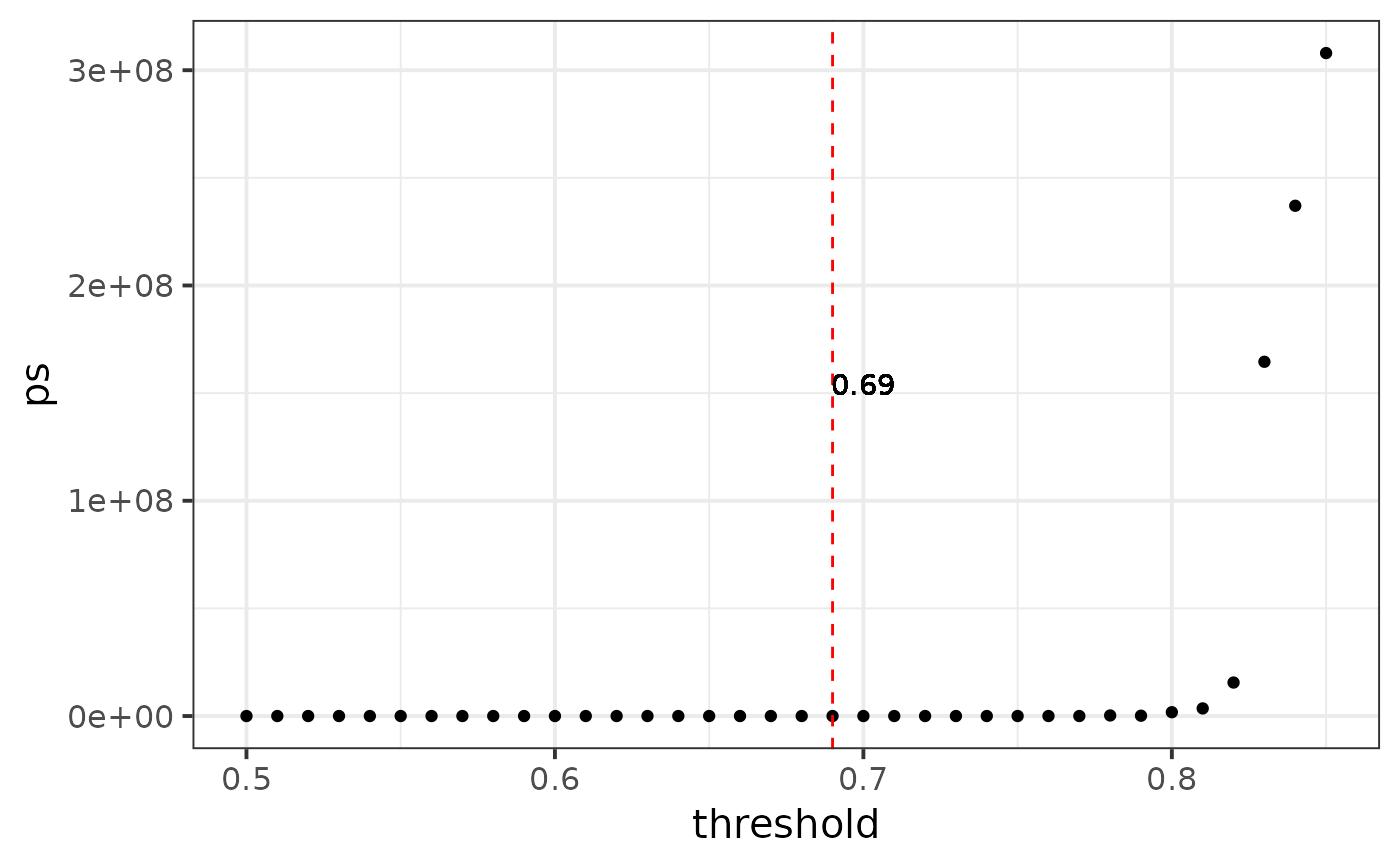
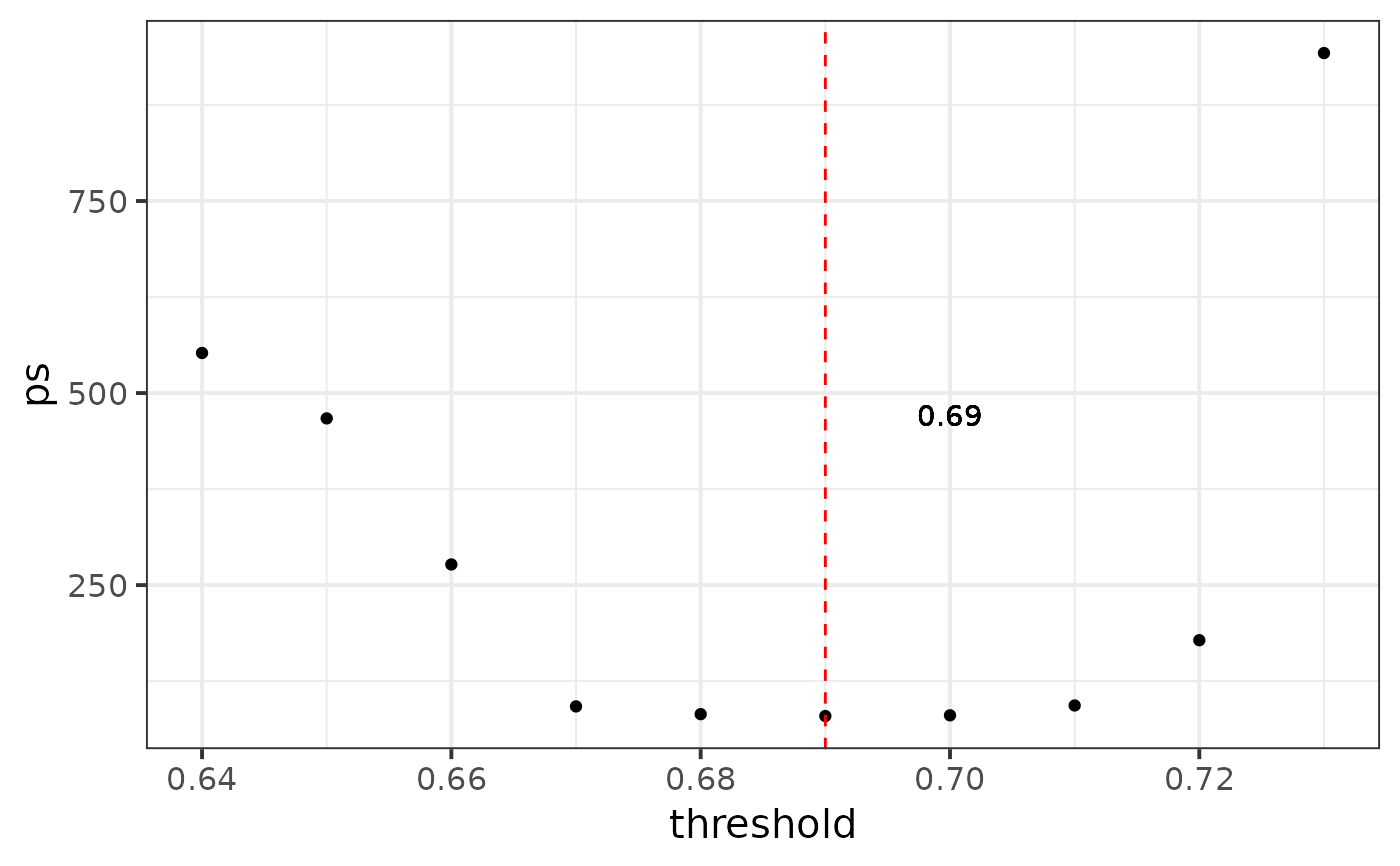 #> We recommend r-threshold: 0.69, you can calculate again in a smaller region
#> [1] 0.69
# recommend: 0.69
c_net_build(corr, r_threshold = 0.69) -> co_net_rmt
#> Have not do p-value adjustment! use the p.value to build network.
# }
#> We recommend r-threshold: 0.69, you can calculate again in a smaller region
#> [1] 0.69
# recommend: 0.69
c_net_build(corr, r_threshold = 0.69) -> co_net_rmt
#> Have not do p-value adjustment! use the p.value to build network.
# }